-Search query
-Search result
Showing 1 - 50 of 103 items for (author: yoshida & m)

EMDB-39463: 
2.6A b-Galactosidase Structure Solved Using an Indirect Scintillator-Coupled CMOS Detector at 300 kV
Method: single particle / : Aramaki S, Yoshida Y, Tanihara T, Oyama K, Otsuki K, Terada Y, Matsunaga N, Ohdo S, Mayanagi K

EMDB-39481: 
2.08A Apoferritin Structure Solved Using an Indirect Scintillator-Coupled CMOS Detector at 300 kV
Method: single particle / : Aramaki S, Yoshida Y, Tanihara T, Oyama K, Otsuki K, Terada Y, Matsunaga N, Ohdo S, Mayanagi K

EMDB-17171: 
CTE typeI tau filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

EMDB-17173: 
CTE typeIII tau filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

EMDB-17174: 
CTE typeII tau filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

EMDB-17175: 
TMEM106B Fold1-s filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

EMDB-17176: 
TMEM106B Fold I-d filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

EMDB-17177: 
Ab typeII filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

EMDB-17178: 
CTE typeI tau filament from Kii ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

EMDB-17179: 
TypeII tau filament from Kii ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

EMDB-17180: 
CTE typeIII tau filament
Method: helical / : Tetter S, Qi C, Ryskeldi-Falcon B, Scheres SHW, Goedert M

EMDB-17181: 
PHF tau filament from Kii ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M
PDB-8ot6: 
CTE typeI tau filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

PDB-8ot9: 
CTE typeIII tau filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M
PDB-8otc: 
CTE typeII tau filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M
PDB-8otd: 
TMEM106B Fold1-s filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

PDB-8ote: 
TMEM106B Fold I-d filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

PDB-8otf: 
Ab typeII filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M
PDB-8otg: 
CTE typeI tau filament from Kii ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M
PDB-8oth: 
TypeII tau filament from Kii ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M
PDB-8oti: 
CTE typeIII tau filament
Method: helical / : Tetter S, Qi C, Ryskeldi-Falcon B, Scheres SHW, Goedert M
PDB-8otj: 
PHF tau filament from Kii ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M

EMDB-16532: 
Structure of Tau filaments Type I from Subacute Sclerosing Panencephalitis
Method: helical / : Qi C, Hasegawa M, Takao M, Sakai M, Akagi M, Iwasaki Y, Yoshida M, Scheres SHW, Goedert M

EMDB-16535: 
Structure of Tau filaments Type II from Subacute Sclerosing Panencephalitis
Method: helical / : Qi C, Hasegawa M, Takao M, Sakai M, Akagi M, Iwasaki Y, Yoshida M, Scheres SHW, Goedert M
PDB-8caq: 
Structure of Tau filaments Type I from Subacute Sclerosing Panencephalitis
Method: helical / : Qi C, Hasegawa M, Takao M, Sakai M, Akagi M, Iwasaki Y, Yoshida M, Scheres SHW, Goedert M

PDB-8cax: 
Structure of Tau filaments Type II from Subacute Sclerosing Panencephalitis
Method: helical / : Qi C, Hasegawa M, Takao M, Sakai M, Akagi M, Iwasaki Y, Yoshida M, Scheres SHW, Goedert M

EMDB-32041: 
Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with short stem
Method: single particle / : Yamada T, Kawamoto A, Yoshida T, Sato Y, Kato T, Tsuge H

EMDB-32043: 
Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem
Method: single particle / : Yamada T, Kawamoto A, Yoshida T, Sato Y, Kato T, Tsuge H

EMDB-33188: 
Complex map of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) di-heptamer
Method: single particle / : Yamada T, Kawamoto A, Yoshida T, Sato Y, Kato T, Tsuge H

EMDB-34136: 
Complex structure of Clostridioides difficile binary toxin folded CDTa-bound CDTb-pore (short).
Method: single particle / : Yamada T, Kawamoto A, Yoshida T, Sato Y, Kato T, Tsuge H

EMDB-34137: 
Complex structure of Clostridioides difficile binary toxin unfolded CDTa-bound CDTb-pore (short).
Method: single particle / : Yamada T, Kawamoto A, Yoshida T, Sato Y, Kato T, Tsuge H

PDB-7vnj: 
Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with short stem
Method: single particle / : Yamada T, Kawamoto A, Yoshida T, Sato Y, Kato T, Tsuge H

PDB-7vnn: 
Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem
Method: single particle / : Yamada T, Kawamoto A, Yoshida T, Sato Y, Kato T, Tsuge H

PDB-7yvq: 
Complex structure of Clostridioides difficile binary toxin folded CDTa-bound CDTb-pore (short).
Method: single particle / : Yamada T, Kawamoto A, Yoshida T, Sato Y, Kato T, Tsuge H

PDB-7yvs: 
Complex structure of Clostridioides difficile binary toxin unfolded CDTa-bound CDTb-pore (short).
Method: single particle / : Yamada T, Kawamoto A, Yoshida T, Sato Y, Kato T, Tsuge H
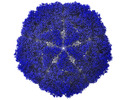
EMDB-12554: 
Bacilladnavirus capsid structure
Method: single particle / : Munke A, Okamoto K

EMDB-12555: 
Genome of an algal ssDNA virus - outer layer
Method: single particle / : Munke A, Okamoto K

EMDB-31526: 
human NTCP in complex with YN69083 Fab
Method: single particle / : Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY

PDB-7fci: 
human NTCP in complex with YN69083 Fab
Method: single particle / : Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY

EMDB-14174: 
TMEM106B filaments with Fold I from Alzheimer's disease (case 1)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW

EMDB-14176: 
TMEM106B filaments with Fold I-d from Multiple system atrophy (case 18)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW

EMDB-14187: 
TMEM106B filaments with Fold IIa from Multiple system atrophy (case 19)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW

EMDB-14188: 
TMEM106B filaments with Fold IIb from Multiple system atrophy (case 19)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW

EMDB-14189: 
TMEM106B filaments with Fold III from Multiple system atrophy (case 17)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW
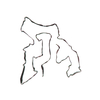
PDB-7qvc: 
TMEM106B filaments with Fold I from Alzheimer's disease (case 1)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW
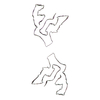
PDB-7qvf: 
TMEM106B filaments with Fold I-d from Multiple system atrophy (case 18)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW
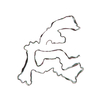
PDB-7qwg: 
TMEM106B filaments with Fold IIa from Multiple system atrophy (case 19)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW
PDB-7qwl: 
TMEM106B filaments with Fold IIb from Multiple system atrophy (case 19)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW

PDB-7qwm: 
TMEM106B filaments with Fold III from Multiple system atrophy (case 17)
Method: helical / : Lovestam S, Schweighauser M, Scheres SHW
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
